|
0.1.13
|
 |
0.1.13
|
| Dependencies | None |
|---|---|
| Python Import | from pyhelios import PhotosynthesisModel |
| Main Class | PhotosynthesisModel |
| Dependencies | None |
|---|---|
| Platforms | Windows, Linux, macOS |
| GPU | Not required |
| Constructors |
|---|
| PhotosynthesisModel |
| Primitive Data Label | Symbol | Units | Data Type | Description | Available Plug-ins | Default Value |
|---|---|---|---|---|---|---|
| radiation_flux_PAR | \(Q\) | W/m2 | float | Radiative flux in PAR band. NOTE: this is automatically converted to units of photon flux density, which are the units used in the photosynthesis model. | Can be computed by RadiationModel plug-in. | 0 |
| temperature | \(T_s\) | Kelvin | float | Primitive surface temperature. | Can be computed by EnergyBalanceModel plug-in. | 300 K |
| air_CO2 | \(C_a\) | \(\mu\)mol CO2/mol air | float | CO2 concentration of air outside of primitive boundary-layer. | N/A | 390 \(\mu\)mol/mol |
| moisture_conductance | \(g_S\) | mol air/m2-s | float | Conductance to moisture between sub-stomatal cells and leaf surface (i.e., stomatal conductance). | Can be computed by StomatalConductanceModel plug-in. | 0.25 mol/m2-s |
| boundarylayer_conductance** | \(g_H\) | mol air/m2-s | float | Conductance to heat between leaf surface and outside of the boundary-layer (i.e., boundary-layer conductance). | Can be computed by BLConductanceModel plug-in, or by EnergyBalanceModel plug-in if optional output primitive data "boundarylayer_conductance_out" is enabled. | 1.0 mol/m2-s |
| twosided_flag | N/A | N/A | uint | Flag indicating the number of primitive faces with heat transfer (twosided_flag = 0 for one-sided heat transfer; twosided_flag = 1 for two-sided heat transfer). This value is retrieved from the primitive's assigned material when available (see Context::getMaterialTwosidedFlag()), with fallback to primitive data if no user-assigned material exists. | N/A | 1 |
| stomatal_sidedness | \(\zeta\) | unitless | float | Ratio of stomatal density on the upper leaf surface to the sum of the stomatal density on upper and lower leaf surfaces. Note: if "twosided_flag" is equal to 0, stomatal_sidedness will be automatically set to 0. | N/A | 0 |
**The photosynthesis model will also check for primitive data "boundarylayer_conductance_out" if "boundarylayer_conductance" does not exist. If you are using the energy balance model to calculate the boundary-layer conductance, you should enable optional output primitive data "boundarylayer_conductance_out" so that other plug-ins can use it.
| Primitive Data Label | Symbol | Units | Data Type | Description |
|---|---|---|---|---|
| net_photosynthesis | \(A\) | \(\mu\)mol CO2/m2-sec | float | Net rate of carbon transfer per unit one-sided area. |
Note: Optional output primitive data functionality is not yet implemented in PyHelios. In native Helios C++, this is done by calling PhotosynthesisModel::optionalOutputPrimitiveData().
| Primitive Data Label | Symbol | Units | Data Type | Description |
|---|---|---|---|---|
| Ci | \(C_i\) | \(\mu\)mol CO2/mol | float | Intercellular CO2 concentration. |
| Gamma_CO2 | \(\Gamma\) | \(\mu\)mol CO2/mol | float | Photosynthetic CO2 compensation point (including "dark respiration"). |
| limitation_state | N/A | N/A | int | Photosynthesis limitation state. limitation_state=0 if photosynthesis is Rubisco-limited, limitation_state=1 if photosynthesis is electron transport limited. |
The PhotosynthesisModel plug-in implements two types of models: the biochemical model of Farquhar, von Caemmerer, and Berry (1980), and an empirical model similar to that of Johnson (2010), which are described separately below.
By default, the plug-in uses the biochemical model. The model can either be set explicitly, as illustrated in the code below, or the model type will be inferred based on the model coefficients that are set (see descriptions below).
The PhotosynthesisModel implements the biochemical model of Farquhar, von Caemmerer, and Berry (1980). The form used here predicts photosynthetic production as a function of photoshynthetically active radiation flux, ambient CO2 concentration, and stomatal conductance, which may itself provide responses to a number of additional environmental variables.
The implementation used here calculates the net rate of CO2 exchange as
\[A=\left(1-\frac{\Gamma^*}{C_i}\right)\mathrm{min}\left\{W_c,W_j,W_p\right\}-R_d,\]
where
\[W_c=\frac{V_{cmax}C_i}{C_i+K_{co}}\]
is the rate limited by Rubisco, with
\[K_{co}=K_c\left(1+\frac{O}{K_O}\right),\]
where \(O\) is oxygen concentration.
\[W_j=\dfrac{J C_i}{4C_i+8\Gamma^*}\]
is the rate limited by RuBP regeneration.
The light response of \(J\), the potential electron transport rate, can be described as a rectangular hyperbola with 1 shape parameter, or non-rectangular hyperbola with 2 shape parameters. The rectangular hyperbola takes the form
\[ J(Q) = \dfrac{J_{max} \alpha Q}{\alpha Q + J_{max}} \]
where \(Q\) is the photosynthetically active absorbed radiation flux ( \(\mu mol\,m^{-2}\,s^{-1}\)), \(J_{max}\) is the temperature-dependent maximum potential electron transport rate ( \(\mu mol\,m^{-2}\,s^{-1}\)), and \(\alpha\) is the intrinsic quantum efficiency of electron transport ( \(electron\,photon^{-1}\)) which defines the initial slope of the light response, determines its resulting shape in the rectangular hyperbolic form, and is thought to be relatively conserved around 0.5. The non-rectangular hyperbola takes the form
\[J(Q) = \dfrac{\alpha Q + J_{max} - \sqrt{(\alpha Q + J_{max})^2 - 4 \theta \alpha Q J_{max}}}{2 \theta}\]
where \(\theta\) is an additional parameter (unitless) that shapes the light response curve beyond its initial slope. When \(\theta\) approaches zero, the two forms become equivalent. In Photosynthesis Plugin, the rectangular form will be assumed unless \(\theta\) is specified by the user.
\[W_p=\dfrac{3\,TPU\,C_i}{C_i-\Gamma^*}\]
is the rate limited by triose phosphate utilization. Note that if the TPU parameter is not set by the user, this state is ignored.
The intercellular CO2 concentration \(C_i\) is determined from the relation
\(A=0.75g_M\left(C_{a}-C_i\right)\)
which is solved numerically using the Secant method, since \(A\) is a complex nonlinear function of \(C_i\) which prevents an analytical solution for \(C_i\). The 0.75 factor comes from the fact that diffusion of CO2 in air is slower than that of water vapor (see Eq. 7.33 of Campbell and Norman).
\(g_M\) is the conductance to moisture transfer between the leaf interior and just outside of the leaf boundary-layer, and is calculated as
\(g_M = 1.08g_Hg_S\left[\dfrac{\zeta}{1.08g_H+g_S\zeta}+\dfrac{(1-\zeta)}{1.08g_H+g_S(1-\zeta)}\right]\),
where \(g_H\) is the boundary-layer conductance to heat, and \(1.08g_H\) gives the boundary-layer conductance to moisture considering the differences in diffusivity between water vapor and heat. \(n_s\) is the number of primitive faces, which is determined by the value of "twosided_flag" retrieved from the primitive's assigned material when available (see Context::getMaterialTwosidedFlag()), with fallback to primitive data if no user-assigned material exists (twosided_flag=0 is single-sided and \(n_s=1\), twosided_flag=1 is two-sided and \(n_s=2\)).
\(\zeta=\dfrac{D_{upper}}{D_{lower}+D_{upper}}\)
is the stomatal sidedness, which is the ratio of the stomatal density of the upper leaf surface to the sum of the upper and lower leaf surface densities, which is set by the primitive data value "stomatal_sidedness". For leaves, \(\zeta=0\) corresponds to hypostomatous leaves (stomata on one side), and \(\zeta=0.5\) to amphistomatous leaves (stomata equally on both sides). It is important to note that if \(n_s=1\), then the value of \(\zeta\) will be overridden and set to 0.
Two different temperature response functions are commonly used in photosynthetic modeling and supported in the Photosynthesis Plugin. One is an Arrhenius equation, which is exponentially increasing with no decline in the region of use. The other is a modified Arrhenius equation with a peak or temperature optimum, beyond which there is a decline in the value of the function, representing a denaturing of an enzyme and subsequent reduction of its activity.
\[ \begin{aligned} k &= k_{25} \cdot \exp \left[\frac{\Delta H_a}{R} \left(\frac{1}{298}-\frac{1}{T_{leaf}} \right) \right] \frac{f(298)}{f(T_{leaf})}, \\ f(T_{leaf}) &= 1+\exp \left[\frac{\Delta H_d}{R} \left(\frac{1}{T_{opt}} - \frac{1}{T_{leaf}} \right) - \ln \left(\frac{\Delta H_d}{\Delta H_a}-1 \right) \right] \end{aligned} \]
In this form, the model is conveniently parameterized by the commonly used standard reference rate at 25 \(^\circ\)C, \(k_{25}\), as well as the energy of activation, \(A = \Delta H_a = dH_a\) of the Arrhenius equation, but also by the observable temperature optimum, \(T_{opt}\) and one additional fitted parameter, the energy of deactivation, \(D = \Delta H _d = dH_d\), related to the rate of decline from the optimum.
As \(T_{opt}\) \(\to \infty\), then the peaked form approaches the standard, unpeaked Arrhenius equation, allowing for mathematical backwards compatibility for parameters obtained from fitting to the standard unpeaked form.
In the Photosynthesis Plugin, the Arrhenius form will be assumed as it requires fewer parameters, unless the additional parameters \(dH_d\) and \(T_{opt}\) are specified by the user.
| Parameter | Description | Units |
|---|---|---|
| \(k_{25}\) | reference rate at 25 \(^\circ\)C | \(\mu mol\,m^{-2}\,s^{-1}\) |
| \(dH_a\) | activation energy (rate of increase) | \(kJ\,mol^{-1}\) |
| \(dH_d\) | deactivation energy (rate of decrease) | \(kJ\,mol^{-1}\) |
| \(T_{opt}\) | optimum temperature in Kelvin | \(K\) |
| \(T_{leaf}\) | leaf surface temperature in Kelvin | \(K\) |
| \(R\) | ideal gas constant, 0.008314 | \(kJ\,mol^{-1}\,K^{-1}\) |
Additional temperature parameters that are not typically fit to use the standard Arrhenius form with the parameters obtained by Bernacchi et al. (2001), and are given by the following equations
\[ \begin{aligned} \Gamma^* &= 42.60 \cdot \exp \left[ \frac{37.83}{R} \left(\frac{1}{298} - \frac{1}{T_{leaf}} \right) \right] \\ K_c &= 400.3 \cdot \exp \left[ \frac{79.43}{R} \left(\frac{1}{298} - \frac{1}{T_{leaf}} \right) \right] \\ K_o &= 275.1 \cdot \exp \left[ \frac{36.38}{R} \left(\frac{1}{298} - \frac{1}{T_{leaf}} \right) \right] \\ R_d &= R_{d,25} \cdot \exp \left[ \frac{46.39}{R} \left(\frac{1}{298} - \frac{1}{T_{leaf}} \right) \right] \end{aligned} \]
| Variable | Units | Description |
|---|---|---|
| \(Q\) | \(\mu\)mol/m2-sec. | Photosynthetic radiation energy flux. |
| \(T_s\) | Kelvin | Surface temperature. |
| \(C_{a}\) | \(\mu\)mol CO2/mol air | Ambient CO2 concentration outside of boundary-layer. |
| \(g_M\) | mol air/m2-s | Conductance to moisture transfer between inside the leaf and leaf surface (i.e., stomatal conductance). |
| \(g_H\) | mol air/m2-s | Conductance to heat transfer between the leaf surface and outside the boundary-layer (i.e., boundary-layer conductance). |
The table below gives example model parameters obtained for several different species. These parameters were fit from leaf-level gas exchange data using the PhoTorch Python package. Note that the parameter sets are based on different temperature response functions depending on the data that was available.
| Species | \(V_{cmax25}\) | \(J_{max25}\) | \(TPU_{25}\) | \(R_{d25}\) | \(\alpha\) | \(\theta\) | \(\Delta H_{a,Vcmax}\) | \(T _{opt,Vcmax}\) | \(\Delta H_{d,Vcmax}\) | \(\Delta H_{a,Jmax}\) | \(T _{opt,Jmax}\) | \(\Delta H_{d,Jmax}\) | \(\Delta H_{a,TPU}\) | \(T _{opt,TPU}\) | \(\Delta H_{d,TPU}\) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Almond | 72.6 | 144.2 | 6.4 | 0.2 | 0.094 | 0 | 27.3 | 315.3 | 478.4 | 64.1 | 314.9 | 508.4 | 37.1 | 311.3 | 477.9 |
| California Bay | 97.5 | 193 | 3.3 | 0.1 | 0.037 | 0 | 49.1 | 308.6 | 505.8 | 34 | 308.5 | 456.7 | 0.1 | 309.4 | 477.5 |
| Elderberry | 37.7 | 149.7 | 7.3 | 1.3 | 0.202 | 0.472 | 66 | 319.4 | 496 | 24.5 | 314.8 | 492.9 | 33.6 | 314.5 | 497.5 |
| Grape | 74.5 | 180.2 | 7.7 | 1.3 | 0.304 | 0 | 76.1 | 318.8 | 499.8 | 23 | 313.8 | 502.3 | 24 | 314.6 | 496.4 |
| Maple | 96.4 | 168 | 2.7 | 0.1 | 0.077 | 0 | 48.9 | 307.1 | 505 | 8.5 | 304.7 | 476.7 | 32.1 | 308.3 | 471.6 |
| Olive | 75.9 | 170.4 | 8.3 | 1.9 | 0.398 | 0 | 55.4 | 315.2 | 497 | 32.2 | 312.5 | 493.4 | 37.2 | 311.7 | 498.9 |
| Pistachio | 101.8 | 223 | 9.8 | 1.5 | 0.216 | 0.65 | 56.5 | 316.6 | 483.1 | 27.7 | 314.6 | 458.5 | 39.9 | 315.4 | 494.3 |
| Toyon | 52.8 | 142.4 | 6.6 | 0.8 | 0.29 | 0.532 | 42.1 | 315.1 | 483 | 9 | 313 | 486.2 | 14 | 314.8 | 493.8 |
| Walnut | 81.6 | 201.9 | 10.2 | 0.9 | 0.362 | 0 | 85.3 | 316.5 | 500.6 | 41.4 | 308.6 | 308.2 | 21.9 | 310.4 | 434.9 |
| Redbud | 68.5 | 132.4 | 6.6 | 0.8 | 0.41 | 0 | 66.6 | 315.1 | 496 | 41.2 | 313.1 | 474 | 34.3 | 312.8 | 463.2 |
| Apple | 101.08 | 167.03 | – | 3.00 | 0.432 | 0 | 65.33 | – | – | 47.62 | – | – | – | – | – |
| Cherry | 75.65 | 129.06 | – | 2.12 | 0.404 | 0 | 65.33 | – | – | 48.49 | – | – | – | – | – |
| Pear | 107.69 | 176.71 | – | 1.51 | 0.274 | 0 | 65.33 | – | – | 46.04 | – | – | – | – | – |
| Prune | 75.88 | 129.41 | – | 1.65 | 0.402 | 0 | 65.33 | – | – | 48.58 | – | – | – | – | – |
The model coefficients can be set manually or by using the library of coefficients provided in the table above.
To load the FvCB parameters from the library, call the function setFarquharCoefficientsFromLibrary() with the species name as an argument. This will automatically set the parameters for all primitives. Alternatively, a list of UUIDs can be passed to this function to set the parameters for a subset of primitives.
To set the parameters manually, first create an instance of the FarquharModelCoefficients class, and then set the parameters using the setter methods. Finally, call the setFarquharModelCoefficients() method with the coefficients object as an argument.
Each parameter has a setter function, which is the means by which the underlying response function to be used is specified.
Parameter Temperature Response
Light Response
Note that the parameter sets in the library vary in terms of which response functions are used based on the data available for parameter fitting.
The native Helios C++ library uses a material-based approach for setting photosynthesis model coefficients:
C++ Approach (Native Helios):
Advantages of material-based approach (C++):
PyHelios Implementation Note:
PyHelios currently uses a UUID-based approach rather than the material-based system. Pass a list of primitive UUIDs to set coefficients for specific primitives:
The UUID-based approach provides similar functionality, allowing you to group primitives logically and set parameters per group.
The PhotosynthesisModel also implements an empirical photosynthesis model using EmpiricalModelCoefficients. The net photosynthetic rate is described by the equation:
\(A = A_{sat} f_L f_T f_C - R_d\)
\(A_{sat}\,({\mu}mol/m^2-s)\) is the photosynthesis assimilation rate at saturating irradiance and reference temperature ( \(T_{ref}\)) and intercellular CO2 concentration ( \(C_{i,ref}\)).
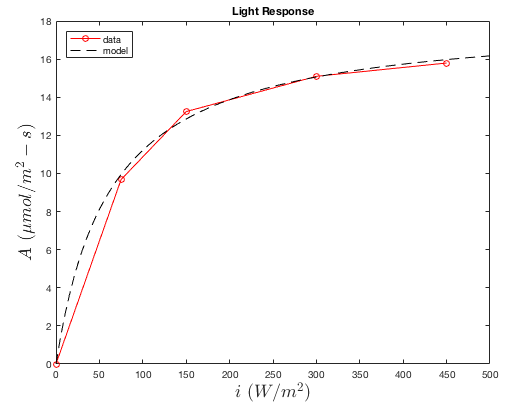
The response of photosynthesis to light is given by a simple exponential function, which is defined by only one parameter:
\(f_L(i) = \dfrac{i}{\theta+i}\),
where \(\theta\) is the light response curvature.
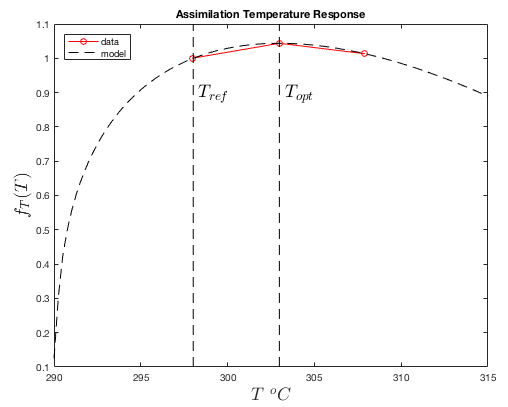
It is assumed that the maximum CO2 assimilation rate \(A_{max}\) decreases exponentially about some optimum temperature \(T_{opt}\). The temperature response function is given by:
\(f_T(T_s) = \left(\dfrac{T_s-T_{min}}{T_{ref}-T_{min}}\right)^q\left(\dfrac{(1+q)T_{opt}-T_{min}-qT_s}{(1+q)T_{opt}-T_{min}-qT_{ref}}\right)\),
where \(T_{min}\) is the minimum temperature at which assimilation occurs, \(T_{opt}\) is the temperature at which the maximum assimilation rate occurs, \(T_{ref}\) is the reference temperature chosen to define \(A_{ref}\), and \(q\) is a shape parameter.
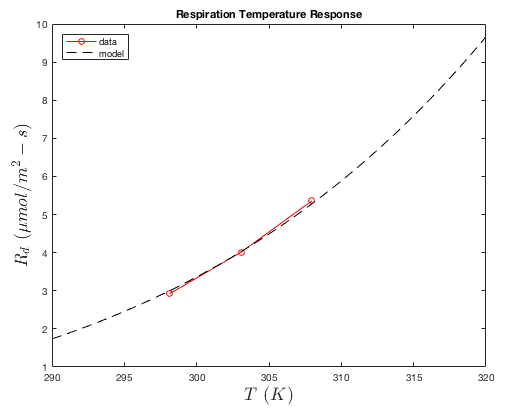
The "dark" respiration rate \(R_d\) is assumed to increase exponentially with temperature following the Arrhenius equation (and assumed not to vary with ambient CO2 concentration). Thus, the dark respiration rate is calculated simply as
\(R_d = R\sqrt{T_s}\mathrm{exp}\left(-E_R/(T_s+273)\right)\),
where \(R\) and \(E_R\) are parameters, and temperature is in Kelvin.
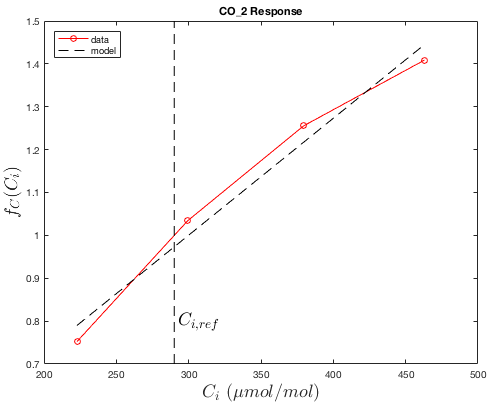
We assume that the maximum assimilation rate varies linearly with intercellular CO2 concentration over the range of expected concentrations, and is zero at zero CO2. Thus, the response function is simply
\(f_C(C_i) = k_C\dfrac{C_i}{C_{i,ref}}\),
where \(C_i\) is intercellular CO2 concentration ( \(\mu\)mol CO2/mol air).
The intercellular CO2 concentration is estimated as a function of the boundary-layer conductance, stomatal conductance, and ambient CO2 concentration outside of the primitive boundary-layer. The rate of transport of CO2 to the leaf (i.e., assimilation rate) is given by
\(A = 0.75g_M\left(C_{amb}-C_i\right)\),
where \(g_M\) is the conductance to moisture from the sub-stomatal cells to outside of the boundary-layer. The 0.75 factor comes from the fact that diffusion of CO2 in air is slower than that of water vapor (see Eq. 7.33 of Campbell and Norman).
Since \(A\) is dependent on \(C_i\) and vice-versa, an iterative solution is required for \(A\).
| Variable | Units | Description |
|---|---|---|
| \(i\) | W/m2 | Photosynthetic radiation energy flux. |
| \(T_s\) | Kelvin | Surface temperature. |
| \(C_{amb}\) | \(\mu\)mol CO2/mol air | Ambient CO2 concentration outside of boundary-layer. |
| \(g_{bl}\) | mol air/m2-s | Boundary-layer conductance. |
| \(g_s\) | mol air/m2-s | Stomatal conductance. |
| Parameter | Units | Description |
|---|---|---|
| \(A_{sat}\) | mol CO2/m2-sec | Assimilation rate at saturating irradiance and reference temperature and intercellular CO2 concentration. |
| \(\theta\) | W/m2 | Shape parameter for response to light. |
| \(T_{min}\) | Kelvin | Minimum temperature at which assimilation occurs. |
| \(T_{opt}\) | Kelvin | Temperature at which maximum assimilation rate occurs. |
| \(q\) | unitless | Temperature response shape function. |
| \(R\) | \({\mu}\)mol K1/2/m2-s | Pre-exponential factor for respiration temperature response. |
| \(E_R\) | 1/Kelvin | Respiration temperature response rate. |
| \(k_C\) | unitless | CO2 response rate. |
The response of the assimilation rate to light is obtained from gas exchange measurements at reference temperature ( \(T_{ref}\)) and CO2 ( \(C_{i,ref}\)) in which the irradiance is varied across some range. However, one important detail is that the dark respiration rate should be removed such that \(A=0\) in the dark (see plot below). This can be done by measuring the net CO2 flux starting in the dark, then subtracting the dark flux from the total flux for each subsequent light level.
The response of the assimilation rate to temperature is obtained using gas exchange measurements at saturating light levels and the reference CO2 concentration. The temperature is varied across some range, and the assimilation rate is measured. It is assumed that the optimum temperature \(T_{opt}\) is the temperature corresponding to the maximum measured assimilation rate. The model is fit to the data to determine \(T_{min}\) and \(q\).
The response of the dark respiration to temperature is obtained using gas exchange measurements in the dark. The leaf is first acclimated to the dark chamber, then leaf temperature is varied across some range. The model is then fit to the data to determine parameters.
The response of the assimilation rate is obtained using gas exchange measurements at saturating light levels and the reference temperature \(T_{ref}\), but with varying external CO2 concentration (which produces varying intercellular CO2).